

FOG01091
EOG8PZGP5
ARO8
sce:ARO8
Genes: 32
EOG8PZGP5
ARO8
sce:ARO8
Genes: 32
Protein description
Aromatic aminotransferase I, anabolic
SGD Description
Aromatic aminotransferase I; expression is regulated by general control of amino acid biosynthesis
PomBase Description
aromatic aminotransferase (predicted)
References
Kradolfer P, et al. (1982 Dec 11). Tryptophan degradation in Saccharomyces cerevisiae: characterization of two aromatic aminotransferases.
Iraqui I, et al. (1998 Jan). Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily.
Urrestarazu A, et al. (1998 Jan). Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination.
Chen D, et al. (2003 Jan). Global transcriptional responses of fission yeast to environmental stress.
Pirkov I, et al. (2008 Aug). A complete inventory of all enzymes in the eukaryotic methionine salvage pathway.
Beltrao P, et al. (2009 Jun 16). Evolution of phosphoregulation: comparison of phosphorylation patterns across yeast species.
Karsten WE, et al. (2011 Dec 1). Mechanism of the aromatic aminotransferase encoded by the Aro8 gene from Saccharomyces cerevisiae.
Van Damme P, et al. (2012 Jul 31). N-terminal acetylome analyses and functional insights of the N-terminal acetyltransferase NatB.
Tarumoto Y, et al. (2013 Jun 28). Receptor for activated C-kinase (RACK1) homolog Cpc2 facilitates the general amino acid control response through Gcn2 kinase in fission yeast.
Carpy A, et al. (2014 Aug). Absolute proteome and phosphoproteome dynamics during the cell cycle of Schizosaccharomyces pombe (Fission Yeast).
Beckley JR, et al. (2015 Dec). A Degenerate Cohort of Yeast Membrane Trafficking DUBs Mediates Cell Polarity and Survival.
Lee J, et al. (2017 Feb 20). Chromatin remodeller Fun30<sup>Fft3</sup> induces nucleosome disassembly to facilitate RNA polymerase II elongation.
| Mitochondrial localization predictions | ||
|---|---|---|
| Predotar | TargetP | MitoProt |
 |
 |
 |
| Raw data
|
||
| Phobius transmembrane predictions
3 genes with posterior transmembrane prediction > 50% |
||
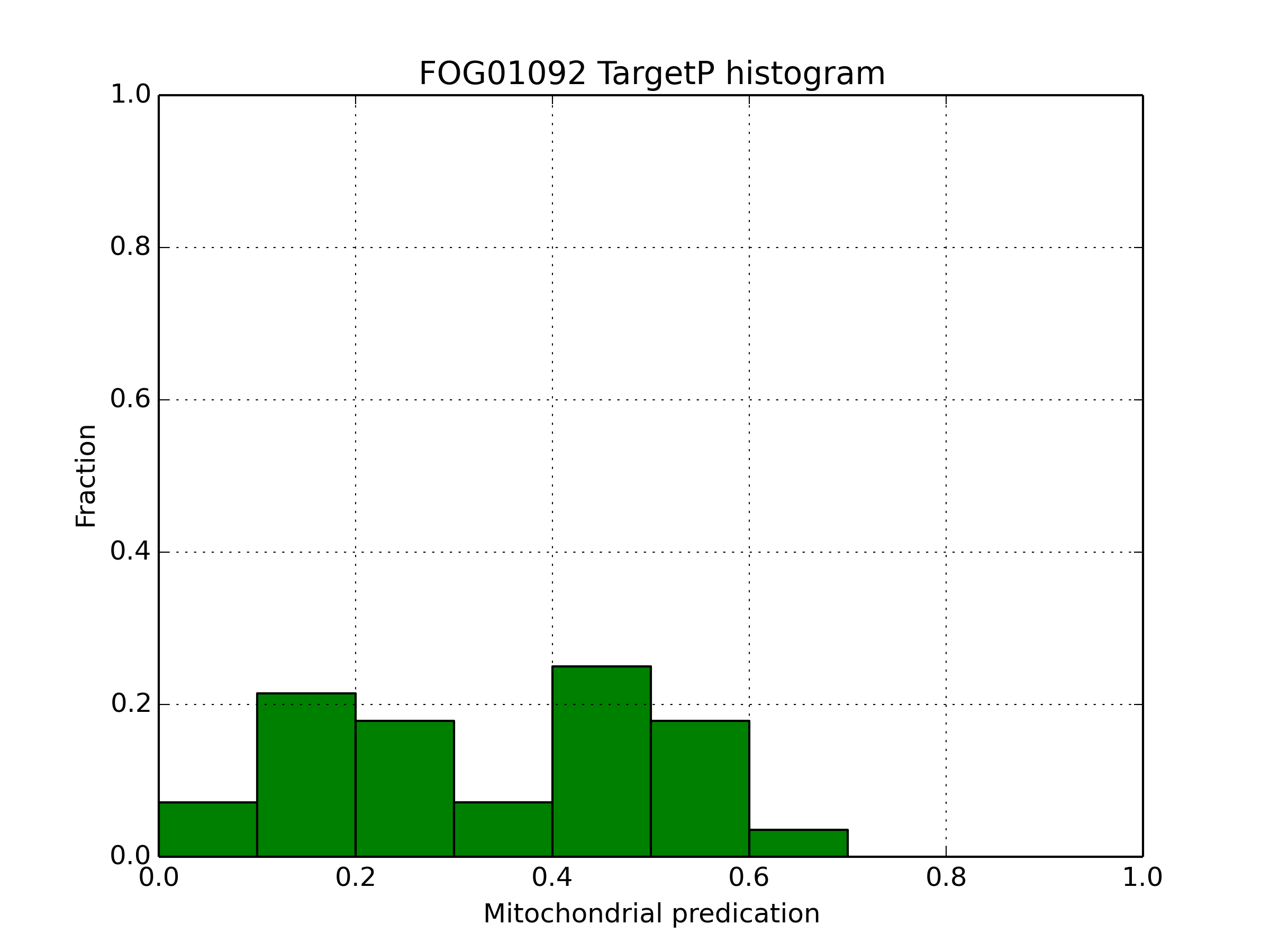
FOG01092
EOG8PZGP5
ARO9
sce:ARO9
Genes: 28
EOG8PZGP5
ARO9
sce:ARO9
Genes: 28
Protein description
Aromatic aminotransferase II, catabolic
SGD Description
Aromatic aminotransferase II; catalyzes the first step of tryptophan, phenylalanine, and tyrosine catabolism
References
Kradolfer P, et al. (1982 Dec 11). Tryptophan degradation in Saccharomyces cerevisiae: characterization of two aromatic aminotransferases.
Iraqui I, et al. (1998 Jan). Characterisation of Saccharomyces cerevisiae ARO8 and ARO9 genes encoding aromatic aminotransferases I and II reveals a new aminotransferase subfamily.
Urrestarazu A, et al. (1998 Jan). Phenylalanine- and tyrosine-auxotrophic mutants of Saccharomyces cerevisiae impaired in transamination.
Iraqui I, et al. (1999 May). Transcriptional induction by aromatic amino acids in Saccharomyces cerevisiae.
Pirkov I, et al. (2008 Aug). A complete inventory of all enzymes in the eukaryotic methionine salvage pathway.
| Mitochondrial localization predictions | ||
|---|---|---|
| Predotar | TargetP | MitoProt |
 |
 |
 |
| Raw data
|
||
| Phobius transmembrane predictions
2 genes with posterior transmembrane prediction > 50% |
||
FOG01093
EOG8PZGP5
sce:absent
Genes: 5
EOG8PZGP5
sce:absent
Genes: 5
AspGD Description
Ortholog(s) have cytosol, nucleus localization
Suggested Analysis
Consolidate with FOG01092?
| Mitochondrial localization predictions | ||
|---|---|---|
| Predotar | TargetP | MitoProt |
 |
 |
 |
| Raw data
|
||
| Phobius transmembrane predictions
0 genes with posterior transmembrane prediction > 50% |
||
FOG01094
EOG8PZGP5
sce:absent
Genes: 3
EOG8PZGP5
sce:absent
Genes: 3
AspGD Description
Has domain(s) with predicted catalytic activity, pyridoxal phosphate binding activity and role in biosynthetic process
Suggested Analysis
Insertion at 300 not present in other homologs; check if monophyletic with YOG00030e; check if arx copy is a xenolog
| Mitochondrial localization predictions | ||
|---|---|---|
| Predotar | TargetP | MitoProt |
 |
 |
 |
| Raw data
|
||
| Phobius transmembrane predictions
1 genes with posterior transmembrane prediction > 50% |
||
FOG01095
EOG8PZGP5
sce:absent
Genes: 3
EOG8PZGP5
sce:absent
Genes: 3
Parent
paralog:FOG01091
| Mitochondrial localization predictions | ||
|---|---|---|
| Predotar | TargetP | MitoProt |
 |
 |
 |
| Raw data
|
||
| Phobius transmembrane predictions
0 genes with posterior transmembrane prediction > 50% |
||
FOG01096
EOG8PZGP5
sce:absent
Genes: 3
EOG8PZGP5
sce:absent
Genes: 3
Parent
paralog:FOG01091
| Mitochondrial localization predictions | ||
|---|---|---|
| Predotar | TargetP | MitoProt |
 |
 |
 |
| Raw data
|
||
| Phobius transmembrane predictions
0 genes with posterior transmembrane prediction > 50% |
||
FOG01097
EOG8PZGP5
sce:absent
Genes: 2
EOG8PZGP5
sce:absent
Genes: 2
AspGD Description
Has domain(s) with predicted catalytic activity, pyridoxal phosphate binding activity and role in biosynthetic process
| Mitochondrial localization predictions | ||
|---|---|---|
| Predotar | TargetP | MitoProt |
 |
 |
 |
| Raw data
|
||
| Phobius transmembrane predictions
0 genes with posterior transmembrane prediction > 50% |
||
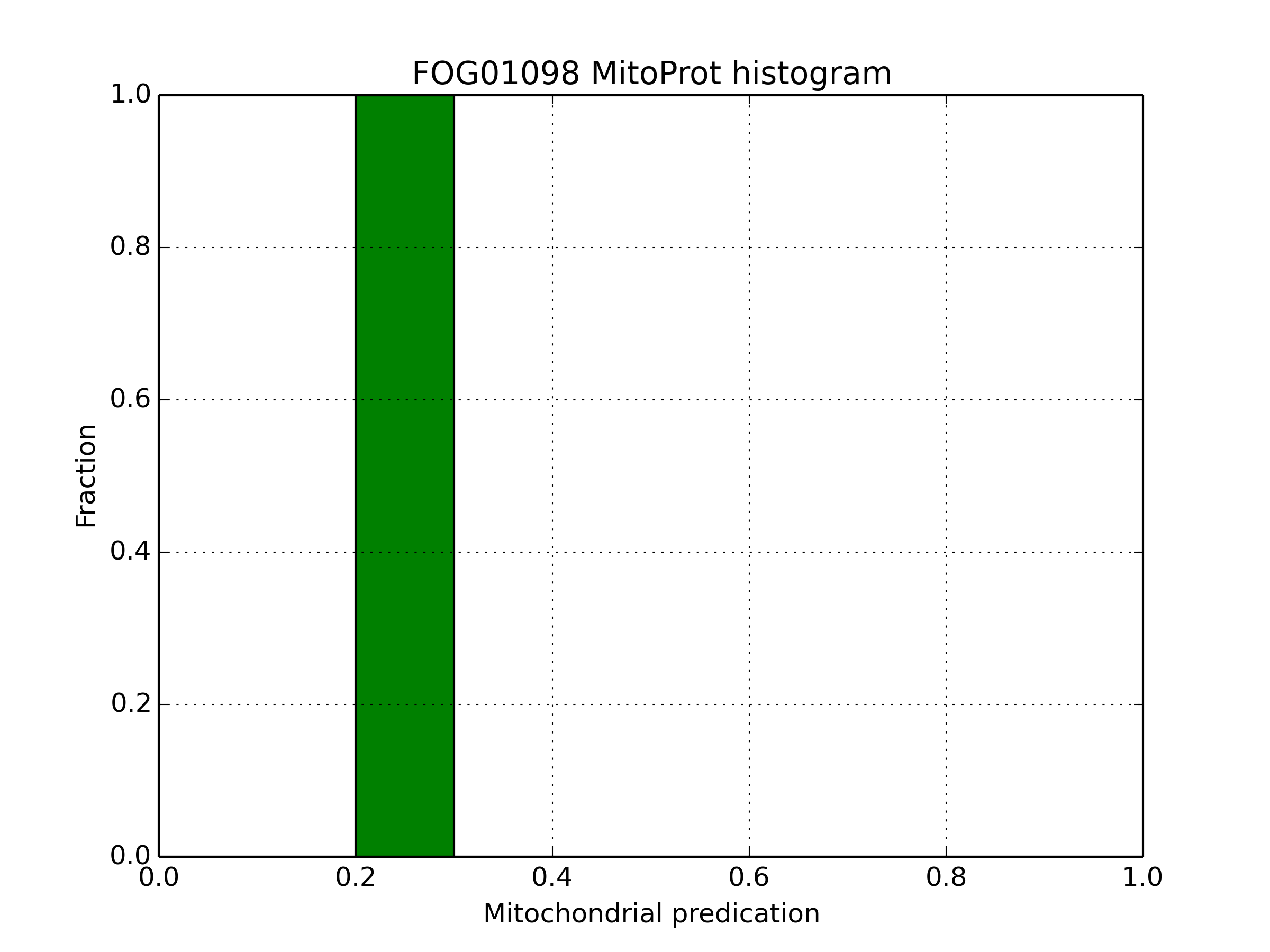
FOG01098
EOG8PZGP5
sce:absent
Genes: 2
EOG8PZGP5
sce:absent
Genes: 2
AspGD Description
Has domain(s) with predicted catalytic activity, pyridoxal phosphate binding activity and role in biosynthetic process
| Mitochondrial localization predictions | ||
|---|---|---|
| Predotar | TargetP | MitoProt |
 |
 |
 |
| Raw data
|
||
| Phobius transmembrane predictions
0 genes with posterior transmembrane prediction > 50% |
||
